[Research] n2v_rc: Node2Vec Recursive Clustering
We constructed a gene co-expression network and developed an algorithm to extract biologically diverse gene groups with graph embedding using node2vec.
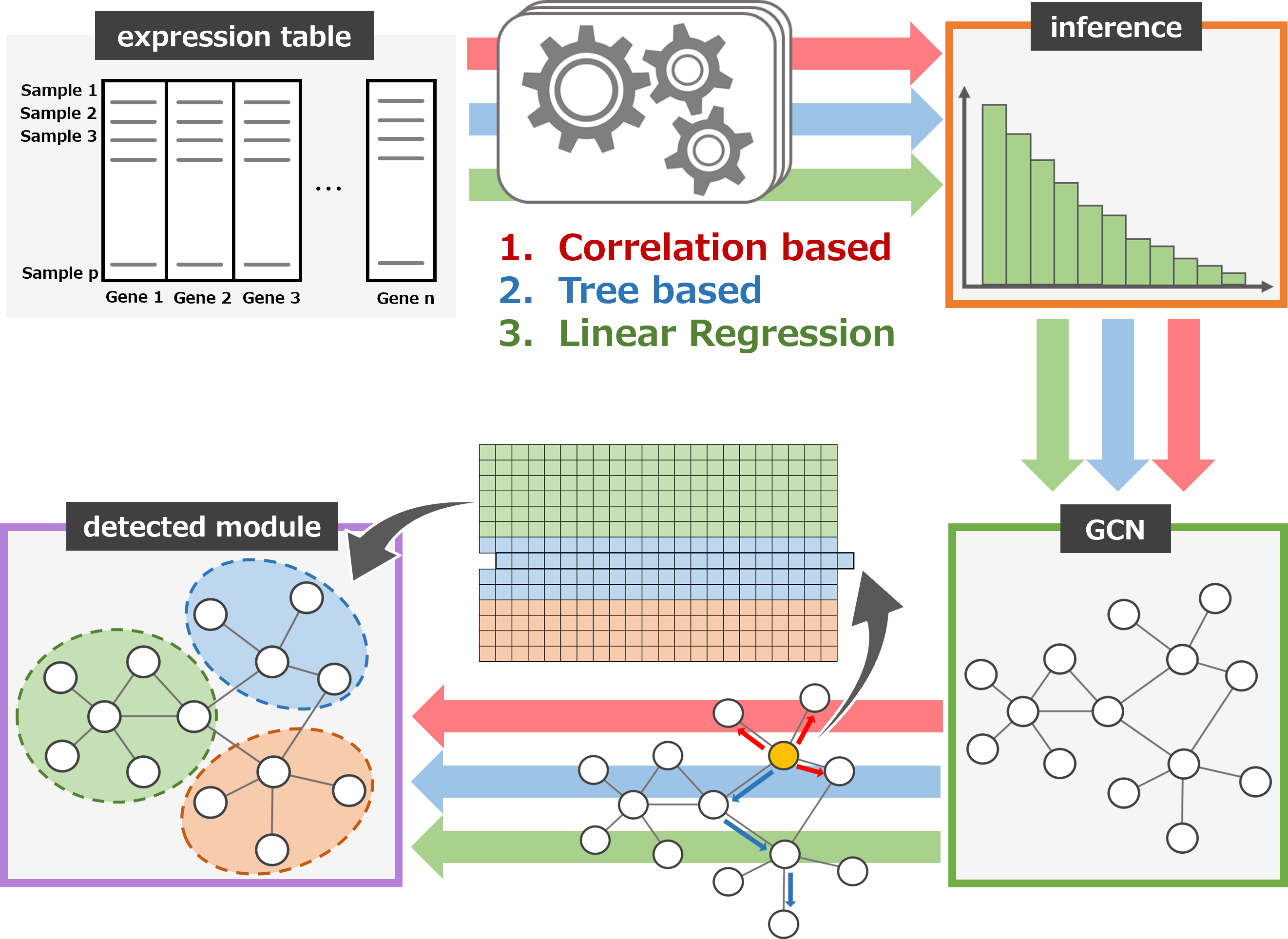
We constructed a gene co-expression network and developed an algorithm to extract biologically diverse gene groups with graph embedding using node2vec.

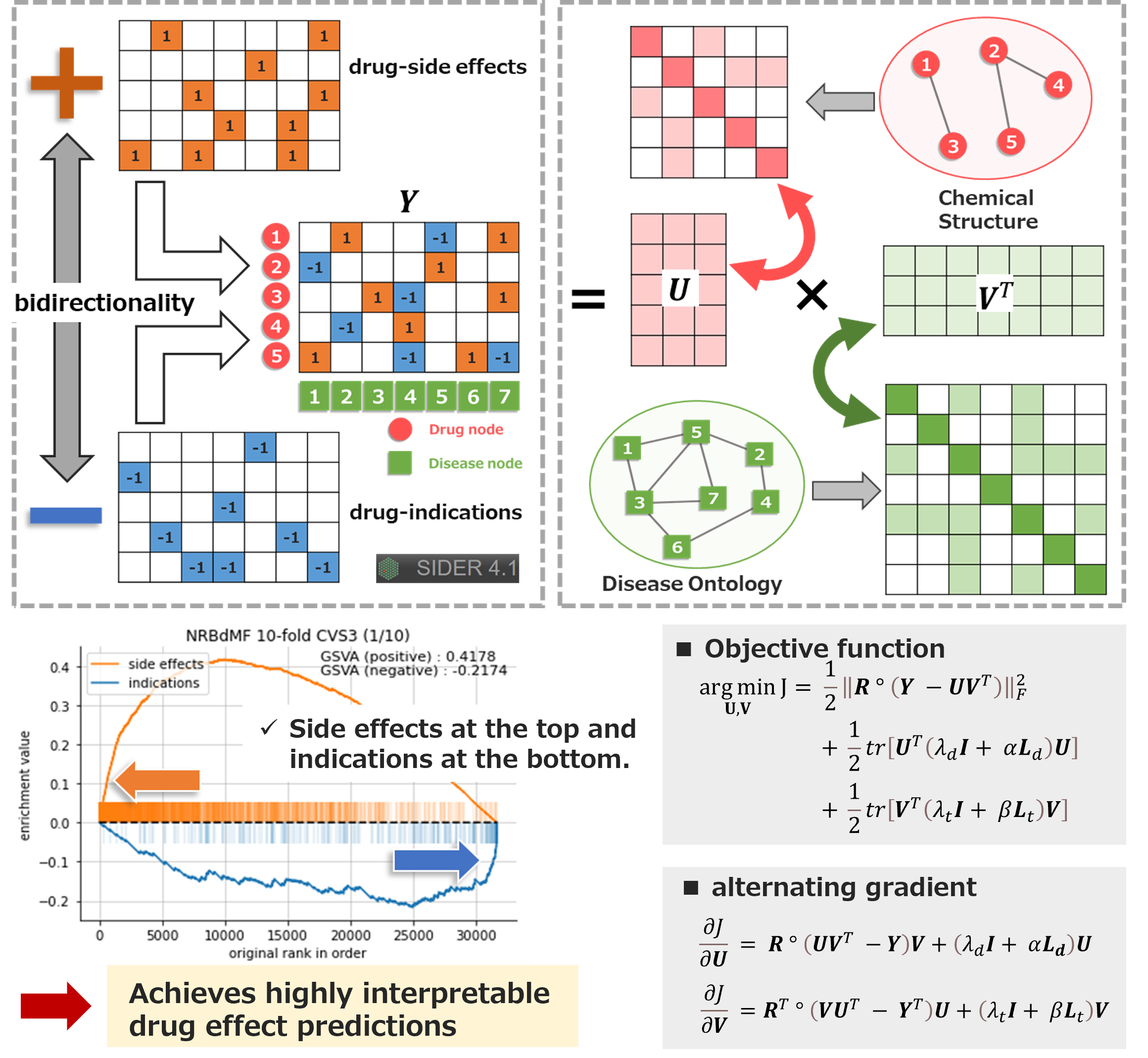
We developed a matrix factorization recommendation algorithm that considers the bidirectional nature of drug effects (side effects - indications) and can provide highly interpretable prediction results.
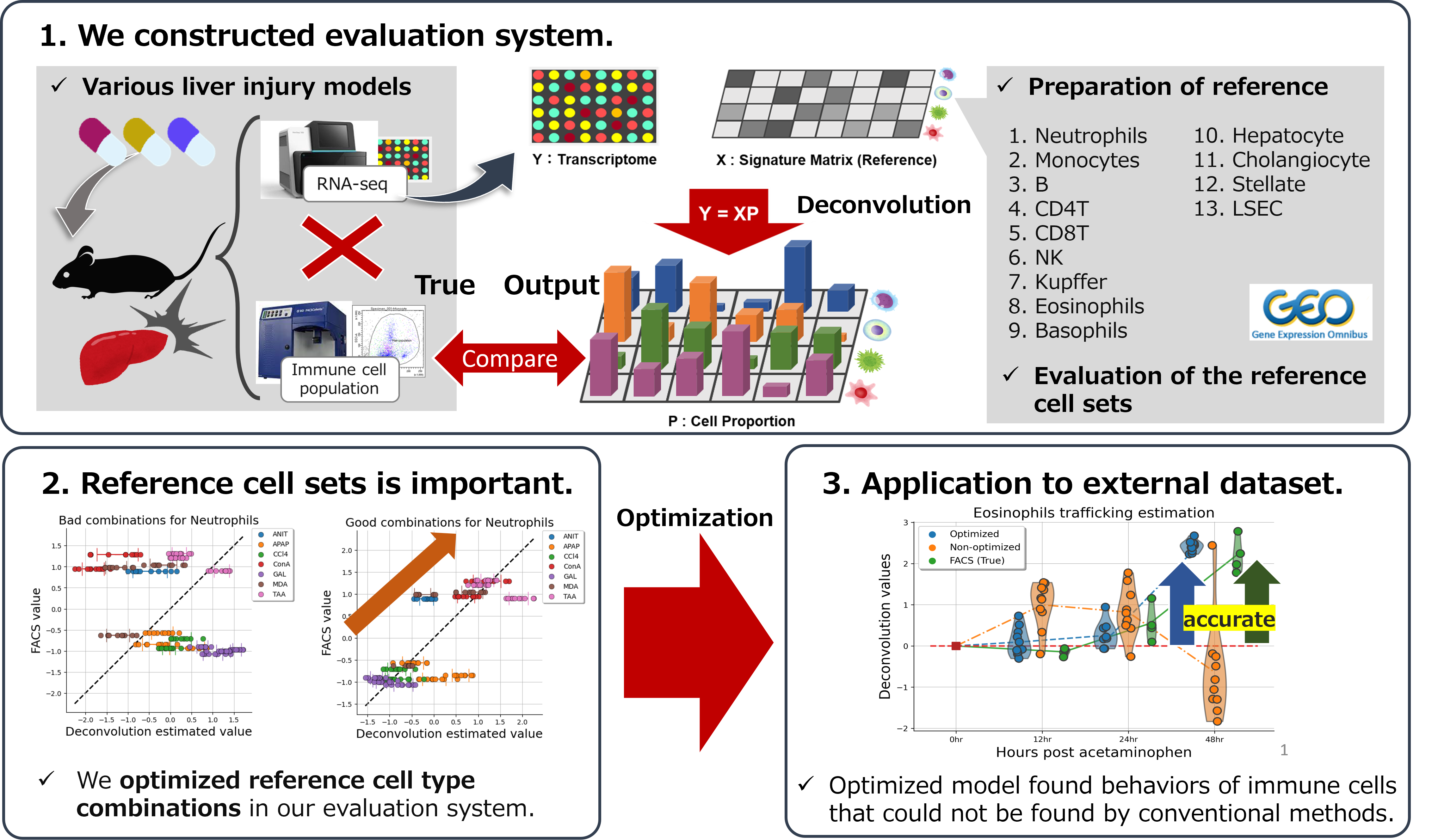
We developed a deconvolution method that can accurately predict the proportions of immune cells from bulk RNA-Seq of liver tissues by modeling the cell types specific to the liver tissue.
「遺伝⼦に関するグラフを利⽤したモデルの開発 」に取り組みました。

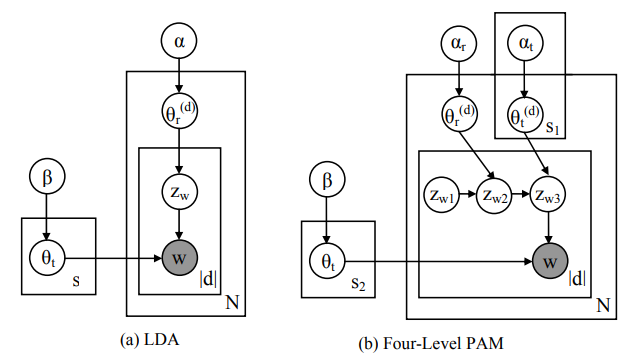
PAMをCythonで実装して高速化しました。
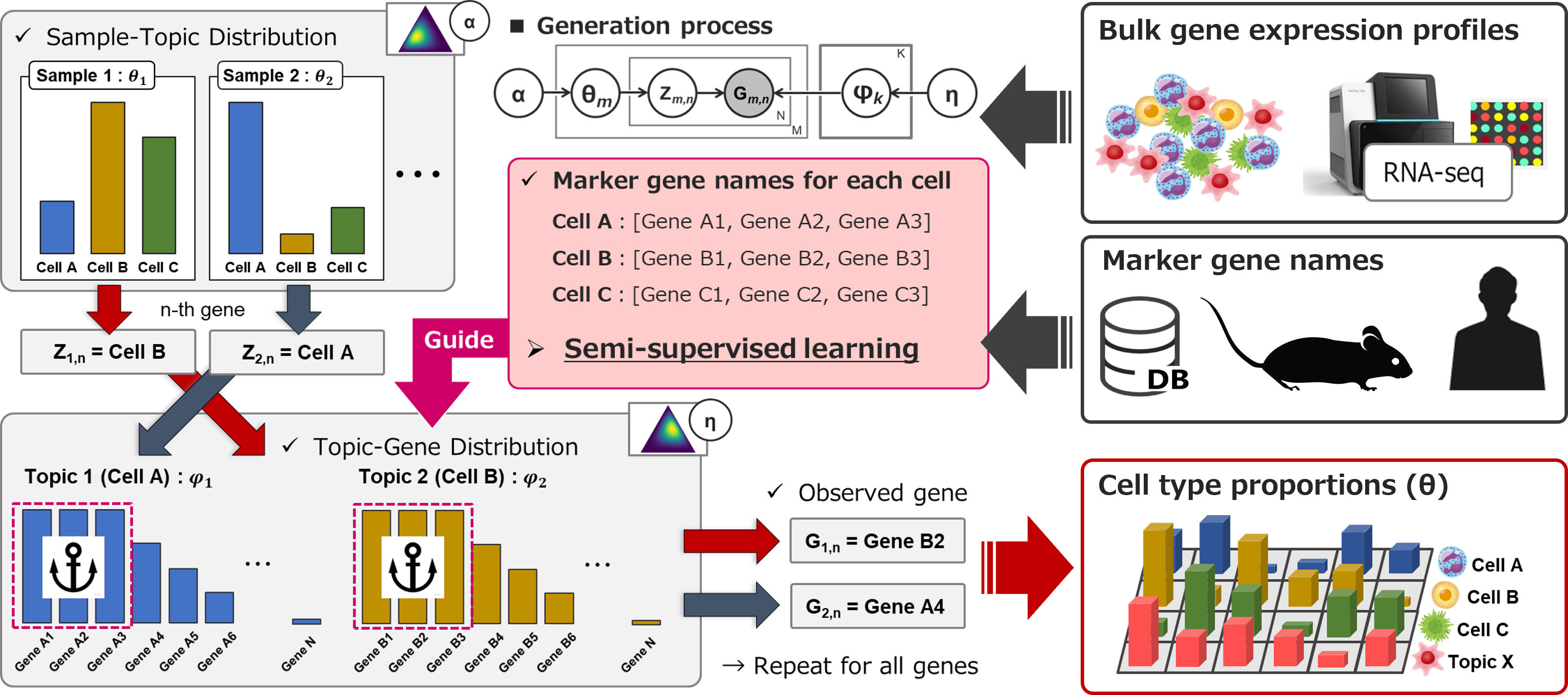
We proposed a Guided LDA Deconvolution method, called GLDADec, to estimate cell type proportions by using marker gene names as partial prior information.